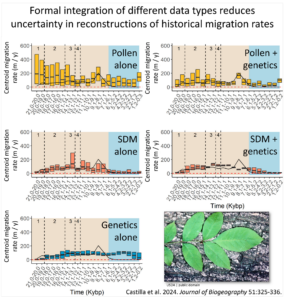
To date, most reconstructions of species’ biogeographic histories have relied on a single data source, such as occurrence data analyzed using species distribution models, fossil pollen analyzed using pollen-vegetation models, or genetics analyzed using scenario-based modeling. “Integration” between them has largely been “by eye.” We present a statistically integrated model based on approximate Bayesian computation (ABC) that can incorporate each of these data types for reconstructing the location of glacial refugia, rate of migration, and population size across time and space.
As an illustration, we analyzed the historical biogeography of Fraxinus pennsylvanica (Green Ash). Including genetic data reduced the uncertainty around estimates of migration rates over time (see figure) and highlighted three general regions where glacial refugia were likely located.
Our software can be applied to a variety of situations to, for example, deduce past pathways of spread of invasive species, evolution and spread of infectious disease, and reconstructing species’ biogeographic histories.
Integrative demographic modeling reduces uncertainty in estimated rates of species’ historical range shifts [open access]
Castilla A.R., A. Brown, S. Hoban, E.A. Abhainn, J.D. Robinson, J. Romero-Severson, A.B. Smith, A.E. Strand, J.R. Tipton, and A. Dawson. 2024. Journal of Biogeography 51:325-336.